Analysing Haemonchus in Tunisian ruminants
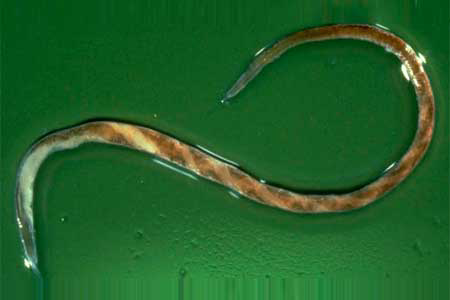
Parasitic gastroenteritis caused by Haemonchus spp. is a major cause of economic losses in the livestock industry, especially in tropical and subtropical areas. The correct identification of various species, as well as knowledge of the epidemiology and genetic characterisation of the principal circulating species, is essential for the establishment of sustainable control strategies.
Haemonchus contortus, also known as red stomach worm or wire worm, is very common parasite and one the most pathogenic nematodes of ruminants. The nematodes piercing the abomasum cause complications in the infected ruminants that can lead to death. They can display severe dehydration, diarrhea, unthrifty appearance, lethargy, depressed low-energy behavior, rough hair coats and uncoordinated movements. Researchers in Tunisia have recently presented epidemiological study of sympatric Haemonchus species and genetic characterisation of Haemonchus contortus in domestic ruminants in Tunisia. It is possibly the first work in North Africa describing the genetic diversity of H. contortus in domestic ruminants.
A study was carried out to determine the prevalence of Haemonchus species in sheep, goats and cattle slaughtered in Béja abattoir from January to June 2010 and also to analyse the genetic differences of Haemonchus contortus in these ruminants. During the study period 364, 271 and 152 abomasa of sheep, goats and cattle respectively, were examined showing overall prevalence rates of 17%, 33.6% and 7.23%, respectively. In addition, spicules morphometric study of 300 male worms randomly collected from sheep showed the presence of 239 (79.66%) H. contortus and 61 (20.33%) H. placei. Likewise, out of 508 adult male Haemonchus from goats, 325 (63.97%) H. contortus and 183 (36.02%) H. placei worms were identified. Whereas for cattle, out of 84 adult male Haemonchus, 52 (61.9%) H. contortus and 32 (38.09%) H. placei worms were identified. The study showed the association of H. contortus and H. placei as a predominant type of infection in all hosts, co-infection concerned 62.5% of sheep, 54.71% of goats and 37.5% of cattle.
Using the polymerase chain reaction, the second Internal Transcribed Spacer region (ITS-2) of the nuclear ribosomal DNA (rDNA) of H. contortus was amplified and sequenced. A total of 16 ITS-2 sequences were identified (five from sheep, three from cattle and eight from goats). The 231 base pairs of different ITS-2 sequences were aligned and analysed. Distance based analysis using Neighbour-Joining method and parsimony analysis were used to construct phylogenetic trees to elucidate genetic relationships. The analyses categorised the ITS-2 sequence of H. contortus into four groups. Groups 1 and 4 were found exclusively in goats, whereas groups 2 and 3 were found in sheep and cattle. This study demonstrates variability in nucleotide sequence within the ITS-2 region that reveals genetic diversity among populations of H. contortus, including those from different domestic ruminant species in Tunisia.
Full report available from ScienceDirect











